New development of biocatalysts
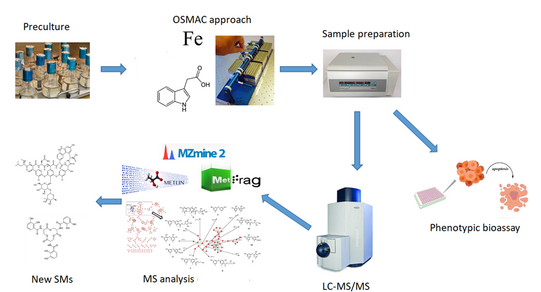
Numerous enzymes and secondary metabolites lie dormant in microorganisms. They can only be discovered with difficulty using traditional screening approaches. Bioprocess technology is therefore working with innovative methods such as genome mining or secondary metabolite screening using the "one strain many compounds" (OSMAC) approach.
Metabolomics
Currently, there is a tremendous need for novel agents for the treatment of diseases and especially to combat multidrug-resistant microbial pathogens. Microbial natural products, especially secondary metabolites, played an important role in the discovery of new drug agents and continue to be a major source of bioactive molecules for drug development. Recently, the discovery of new bioactive natural products has slowed down due to the silencing of the formation of most secondary metabolites encoded by biosynthetic gene clusters under standard cultivation conditions and the immense rediscovery of already known products under the time and labor intensive traditional screening methods [1].
In recent decades, various methods such as ribosome engineering, heterologous expression, manipulation or regulation of promoters, induction of mutations, quorum sensing and elicitor screening systems, co-cultivation, and altered cultivation conditions have been introduced to activate silent biosynthetic gene clusters and increase the chance of discovering new bioactive SMs [2]. Manipulation of cultivation conditions has been reported in the literature as the most effective approach, referred to as the OSMAC (one strain many compounds) strategy. OSMAC describes the production of many SMs from one microorganism by changing the culture conditions such as temperature, pH, oxygenation, culture vessel, media composition, addition of enzyme inhibitors and small molecules [3]. In this project, we aim to discover new secondary metabolites and activation of silent gene clusters in bacteria by applying metabolomics screening strategies using the "one strain many compounds" approach.
[1] Katz, L. and Baltz, R., 2016. Natural product discovery: past, present, and future. Journal of Industrial Microbiology & Biotechnology, 43(2-3), pp.155-176. [2] Zarins-Tutt, J., Barberi, T., Gao, H., Mearns-Spragg, A., Zhang, L., Newman, D. and Goss, R., 2016. Prospecting for new bacterial metabolites: a glossary of approaches for inducing, activating and upregulating the biosynthesis of bacterial cryptic or silent natural products. Natural Product Reports, 33(1), pp.54-72. [3] Pan, R., Bai, X., Chen, J., Zhang, H. and Wang, H., 2019. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Frontiers in Microbiology, 294(10).
Contact
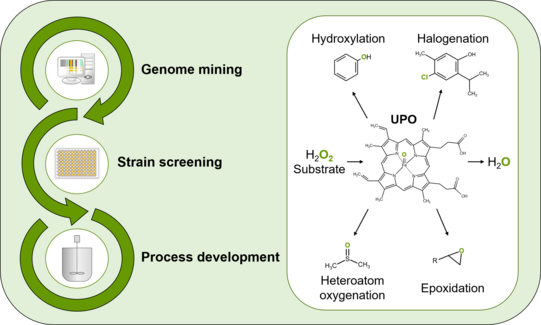
Genome mining and enzyme screening
In 2004, a new heme thiolate peroxidase was discovered in the basidiomycete Agrocybe aegerita that naturally catalyzes a broad spectrum of oxidative transformations [1].
This novel enzyme type, classified as unspecific peroxygenases (UPOs, EC 1.11.2.1), is found throughout the fungal kingdom and combines the catalytic cycle of heme peroxidases with the "peroxide shunt" of cytochrome P450 monooxygenases (CYPs). In general, promiscuous biocatalysts transfer one oxygen atom from hydrogen peroxide to numerous substrates and are therefore particularly promising for applications involving the selective oxyfunctionalization of organic molecules [2].
Due to the continuous demand for new biocatalysts with novel reaction and substrate spectra, it is of great interest to identify and characterize putative UPOs. In this project, a combination of bioinformatics analysis and enzyme activity screening is used to identify rarely observed fungal strains and their UPOs as promising biocatalysts for hydroxylation reactions. Further optimization of wild-type strains as well as heterologously expressed UPOs may lead to the development of new oxyfunctionalization processes for biotechnological applications.
[1] R. Ullrich, J. Nueske, K. Scheibner, J. Spantzel, and M. Hofrichter, "Novel haloperoxidase from the Agaric Basidiomycete," Appl. Environ. Microbiol. vol. 70, no. 8, pp. 4575-4581, 2004.
[2] M. Hofrichter, H. Kellner, R. Herzog, A. Karich, C. Liers, K. Scheibner, V. Wambui Kimani, and R. Ullrich, "Fungal Peroxygenases: A Phylogenetically Old Superfamily of Heme Enzymes with Promiscuity for Oxygen Transfer Reactions," in Grand Challenges in Fungal Biotechnology, P. H. Rampelotto, Springer, pp. 369-403, 2020.
Contact
Alina Kinner
Cyclic dinucleotides
In the presence of cytosolic DNA, the enzyme cyclic GMP-AMP synthase (cGAS) catalyzes the synthesis of the second messenger 2'3'-cyclic GMP-AMP (cGAMP) [1].
The cyclic dinucleotide cGAMP is currently of great interest to the pharmaceutical industry because it can activate the innate immune system [2]. Potential applications include immunotherapy of cancer or use as an adjuvant in vaccines [3]. The aim of this PhD topic is to develop an enzymatic process for the production of the relevant cyclic dinucleotides.
[1] Sun, L., et al, Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science (New York, N.Y.), 2013. 339(6121): p. 786-791. [2] Corrales, L., et al, Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell reports, 2015. 11(7): p. 1018-1030. [3] Cai, X., Y.-H. Chiu, and Z.J. Chen, The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Molecular cell, 2014. 54(2): p. 289-296.
Contact
Martin Becker







![[Translate to English:] [Translate to English:]](/storages/bpt-bci/_processed_/0/d/csm_Projektabbildung_Martin_fde7b13e81.jpg)